메뉴
KR | KRW
KR | KRW

How do I spend less time on PCR experiments and get results faster?
실험실 아카데미
- 제약
- 효율성
- 증폭 및 PCR
- PCR Cycler
- 논문
Much time in the laboratory is spent on conducting PCR experiments. It is worth analyzing which aspects contribute to the total time spent, and where effort can be reduced. One obvious point is pure run time, but attention should also be paid to the preparation of the experiments, as well as to the question of whether PCR runs could possibly be avoided altogether.
Speed up your reaction – Part 1: Minimization of reaction time
A PCR run (traditionally) consists of three temperature steps (denaturation, annealing and elongation) that run in cycles (figure 1). Certain parameters are dependent on the type and the amount of template as well as on the primers used, but there are many parameters that are amenable to “tweaking” when it comes to shortening the reaction time and to performing a “fast PCR”. 자세히 보기
간단히 보기
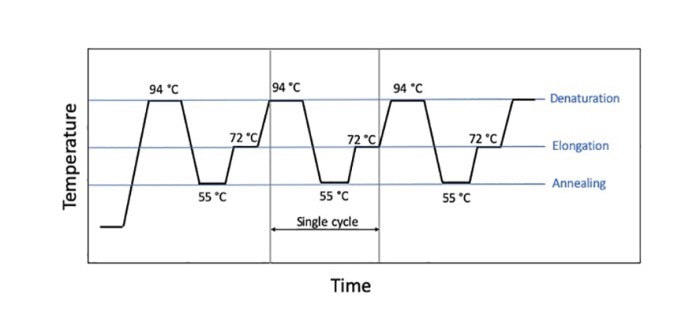
Figure 1: Sequence of a classic 3-step PCR
On the one hand, it is important that the reaction components hold the target temperatures of the cycles for a certain period of time in order to ensure an optimal, i.e. specific, PCR run with high yield. For example, the duration of the denaturation step depends on the GC-content and possible secondary structures of the target sequence, and the elongation time depends on the length of the target sequence. The number of cycles, on the other hand, is dictated by the copy number which enters into the reaction. Specific PCR reagents such as, for example, a modified polymerase that displays higher processivity, or additives that accelerate primer annealing or influence the melting behavior of the DNA, respectively, may help reduce the holding times required, and thus the duration of the run, especially in the case of long amplicons.
Since PCR consists of a large number of temperature changes, the speed at which the sample reaches the required temperature is also relevant. This speed is determined by three main factors: the thermocycler, the consumables and the sample volume. The speed is a direct result of the active heating and cooling capacity of the instrument in combination with the conductivity of the heating block. Silver, for example, displays higher thermal conductivity than aluminum (table 1), which affects the heating and cooling rates of cyclers with blocks made from the respective materials (figure 2).
On the one hand, it is important that the reaction components hold the target temperatures of the cycles for a certain period of time in order to ensure an optimal, i.e. specific, PCR run with high yield. For example, the duration of the denaturation step depends on the GC-content and possible secondary structures of the target sequence, and the elongation time depends on the length of the target sequence. The number of cycles, on the other hand, is dictated by the copy number which enters into the reaction. Specific PCR reagents such as, for example, a modified polymerase that displays higher processivity, or additives that accelerate primer annealing or influence the melting behavior of the DNA, respectively, may help reduce the holding times required, and thus the duration of the run, especially in the case of long amplicons.
Since PCR consists of a large number of temperature changes, the speed at which the sample reaches the required temperature is also relevant. This speed is determined by three main factors: the thermocycler, the consumables and the sample volume. The speed is a direct result of the active heating and cooling capacity of the instrument in combination with the conductivity of the heating block. Silver, for example, displays higher thermal conductivity than aluminum (table 1), which affects the heating and cooling rates of cyclers with blocks made from the respective materials (figure 2).
Since PCR consists of a large number of temperature changes, the speed at which the sample reaches the required temperature is also relevant. This speed is determined by three main factors: the thermocycler, the consumables and the sample volume. The speed is a direct result of the active heating and cooling capacity of the instrument in combination with the conductivity of the heating block. Silver, for example, displays higher thermal conductivity than aluminum (table 1), which affects the heating and cooling rates of cyclers with blocks made from the respective materials (figure 2).
On the one hand, it is important that the reaction components hold the target temperatures of the cycles for a certain period of time in order to ensure an optimal, i.e. specific, PCR run with high yield. For example, the duration of the denaturation step depends on the GC-content and possible secondary structures of the target sequence, and the elongation time depends on the length of the target sequence. The number of cycles, on the other hand, is dictated by the copy number which enters into the reaction. Specific PCR reagents such as, for example, a modified polymerase that displays higher processivity, or additives that accelerate primer annealing or influence the melting behavior of the DNA, respectively, may help reduce the holding times required, and thus the duration of the run, especially in the case of long amplicons.
Since PCR consists of a large number of temperature changes, the speed at which the sample reaches the required temperature is also relevant. This speed is determined by three main factors: the thermocycler, the consumables and the sample volume. The speed is a direct result of the active heating and cooling capacity of the instrument in combination with the conductivity of the heating block. Silver, for example, displays higher thermal conductivity than aluminum (table 1), which affects the heating and cooling rates of cyclers with blocks made from the respective materials (figure 2).
자세히 보기
간단히 보기

Thermal conductivity at 20 °C: Aluminium: 98.8 [mm2/s] | Silver: 173 [mm2/s]
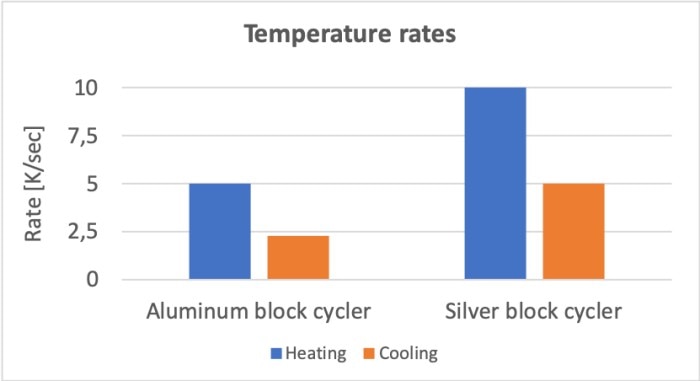
Figure 2: Comparison of heating and cooling rates between a cycler with an aluminum block (Mastercycler® X50a) and a cycler with a silver block (Mastercycler® X50s).
The heating and cooling rates of an instrument are generally published citing their technical data. These, however, do not always accurately represent a direct and therefore comparable measure of their actual speed as they depend on the way in which were calculated. This fact is clearly illustrated in Application Note 274, in which two cycler variants which both display a ramp rate of 5 °C (according to their technical data) nevertheless differ in their total run times by more than 10 minutes [1], as well as through forensic DNA typing that was carried out using six different types of cyclers. This example, too, shows that the published ramp rates do not necessarily have to correlate with the actual run times [2]. In fact, comparing the actual run times of a single protocol executed on different cyclers is well worth the effort.
Achieving rapid temperature changes of the block is the basic prerequisite – the actual goal, however, is reaching the respective temperature inside the sample. Between heating block and sample, there is the vessel. In order to ensure optimized temperature transition, the shape of the vessel should fit the shape of the cycler block perfectly, and its wall should be as thin as possible. Standard PCR Consumables consist of polypropylene. This material offers many advantages; sufficiently fast temperature transfer, however, can only be achieved with thin vessel walls (figure 3).
Achieving rapid temperature changes of the block is the basic prerequisite – the actual goal, however, is reaching the respective temperature inside the sample. Between heating block and sample, there is the vessel. In order to ensure optimized temperature transition, the shape of the vessel should fit the shape of the cycler block perfectly, and its wall should be as thin as possible. Standard PCR Consumables consist of polypropylene. This material offers many advantages; sufficiently fast temperature transfer, however, can only be achieved with thin vessel walls (figure 3).
자세히 보기
간단히 보기
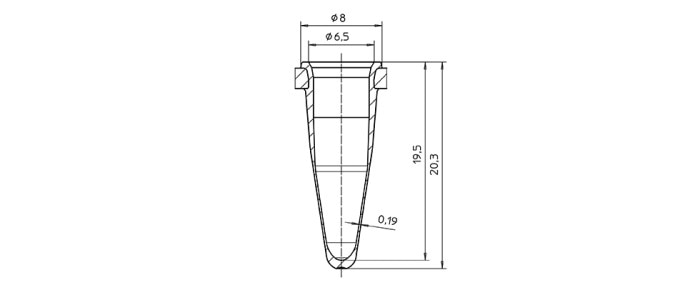
Figure 3: Graphic representation of a well of a Eppendorf twin.tec® PCR plate with a very thin wall to ensure good temperature transfer.
Consumables made of polyethylene represent a viable alternative. This material possesses higher heat conductivity than polypropylene. As shown in Application Note 400, these consumables make it possible to turn a standard PCR protocol directly into a fast PCR protocol without compromising yield and without the need for optimizing the reaction [3].
A further option capable of shortening heating and cooling times of the sample, respectively, is a reduction in sample volume. If large sample numbers are processed simultaneously, 384 well plates are a good choice. If work is performed in the 96-well format, plates and vessels of the “low profile” versions lend themselves particularly well to the use of small volumes (figure 4). The area of the vessel wall between sample and heated lid is reduced in “low profile” versions, thus generating less condensation that would change the concentration of the reaction components.
A further option capable of shortening heating and cooling times of the sample, respectively, is a reduction in sample volume. If large sample numbers are processed simultaneously, 384 well plates are a good choice. If work is performed in the 96-well format, plates and vessels of the “low profile” versions lend themselves particularly well to the use of small volumes (figure 4). The area of the vessel wall between sample and heated lid is reduced in “low profile” versions, thus generating less condensation that would change the concentration of the reaction components.
자세히 보기
간단히 보기
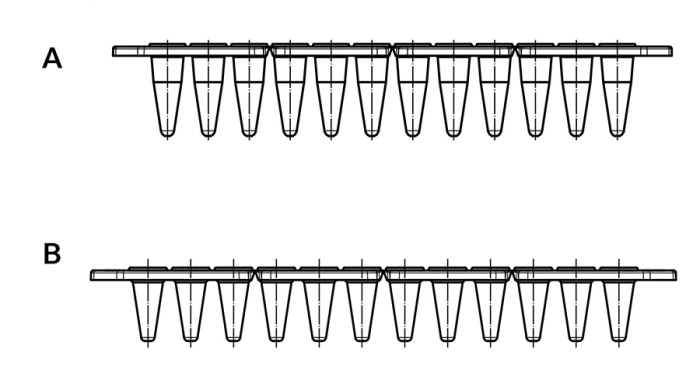
Figure 4:
A Eppendorf twin.tec®, unskirted low profile 150 µL
B Eppendorf twin.tec®, unskirted 250 µL
A Eppendorf twin.tec®, unskirted low profile 150 µL
B Eppendorf twin.tec®, unskirted 250 µL
Small reaction volumes, however, also have disadvantages. Manual pipetting of reactions of such small volumes, especially, can prove challenging; it is prone to error and it can lead to incorrect results. It is therefore beneficial to automate the preparation of very small reaction volumes in order to ensure accurate dosing and thus avoid possible errors. At the same time, a tight seal of the vessel needs to be in place in order to minimize evaporation. In this way, for example, a “low profile” PCR plate will allow a successful PCR to be performed in a volume as low as 3 µL [4].
A different way to shorten the reaction time involves the conversion of the classic three-step PCR to a 2-step PCR (figure 5). In this case, annealing and elongation occur in the same step, meaning that one step and one temperature change are saved in each cycle. Many experiments permit this method as most polymerases are sufficiently active at the chosen annealing temperature.
A different way to shorten the reaction time involves the conversion of the classic three-step PCR to a 2-step PCR (figure 5). In this case, annealing and elongation occur in the same step, meaning that one step and one temperature change are saved in each cycle. Many experiments permit this method as most polymerases are sufficiently active at the chosen annealing temperature.
자세히 보기
간단히 보기
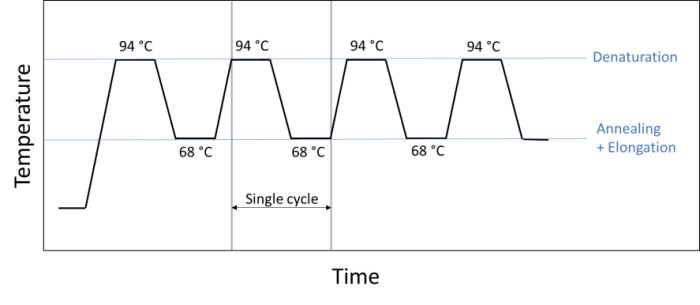
Figure 5: Sequence of a 2-step PCR during which annealing and elongation are carried out in a single step (e.g. at 68 °C).
The combination of a fast instrument, fast PCR reagents, thin-walled consumables and a switch to two-step PCR can help shorten run times considerably and thus expedite the workflow in the laboratory.
Read all about it in the next part: “Speed up your reaction – Part 2: Reduction of preparation time and avoiding runs” shows how the effort involved in the preparation of a PCR can be reduced and which strategies exist that can help decrease the number of PCR runs that need to be performed.
Read all about it in the next part: “Speed up your reaction – Part 2: Reduction of preparation time and avoiding runs” shows how the effort involved in the preparation of a PCR can be reduced and which strategies exist that can help decrease the number of PCR runs that need to be performed.
자세히 보기
간단히 보기
References:
[1] Eppendorf Application Note 274
[2] Butts ELR, Vallone PM. Rapid PCR protocols for forensic DNA typing on six thermal cycling platforms. Electrophoresis. 2014; 35: 3053-3061. Butts EL1, Vallone PM.
[3] Eppendorf Application Note 400
[4] Eppendorf Application Note 388
Eppendorf® , the Eppendorf Brand Design, Eppendorf twin.tec® and Mastercycler® are registered trademarks of Eppendorf AG, Germany. All rights reserved, including graphics and images · Copyright © 2020 by Eppendorf AG.
[1] Eppendorf Application Note 274
[2] Butts ELR, Vallone PM. Rapid PCR protocols for forensic DNA typing on six thermal cycling platforms. Electrophoresis. 2014; 35: 3053-3061. Butts EL1, Vallone PM.
[3] Eppendorf Application Note 400
[4] Eppendorf Application Note 388
Eppendorf® , the Eppendorf Brand Design, Eppendorf twin.tec® and Mastercycler® are registered trademarks of Eppendorf AG, Germany. All rights reserved, including graphics and images · Copyright © 2020 by Eppendorf AG.
자세히 보기
간단히 보기

